Behrens-Fisher
Freddy Hernandez and Jean Paul Piedrahita
2025-09-11
Source:vignettes/Behrens-Fisher.Rmd
Behrens-Fisher.RmdSummary
In this vignette, we introduce the stests R package, which serves as
a valuable tool for testing hypotheses related to the Berens-Fisher
problem. The package is conveniently hosted on GitHub and is freely
accessible for utilization. The core functionality of the stests package
revolves around the two_mean_vector_test function. This
function generates a specialized R object that allows the use of the
standard print function, enabling easy examination of the test results.
Furthermore, the two_mean_vector_test function offers the
flexibility to generate informative plots showcasing the p-value
alongside the corresponding statistics and probabilities. These
visualizations provide a comprehensive understanding of the hypothesis
testing process, enhancing the interpretability and usability of the
stests package. Researchers and practitioners alike can leverage this
package to conduct hypothesis testing for the Berens-Fisher problem
effectively and efficiently.
Introduction
The multivariate Behrens-Fisher problem deals with testing whether two mean vectors are equal or not based on two random samples obtained from two multivariate normal populations. Symbolically, the Behrens-Fisher problem can be summarized as follows.
Let a random sample from a -variate normal population with . The Behrens-Fisher problem has the next set of hypothesis.
The covariance matrices can be unknown and possibly unequal.
Multiple solutions have been proposed to tackle the multivariate Behrens-Fisher problem. The two earliest solutions were given by Bennett (1951) based on the univariate solution of Scheffé (1943) and by James (1954) who extended the univariate Welch series solution.
Yao (1965) proposed an extension of the Welch approximate degrees of freedom and performed a simulation study to compare it with the James solution.
Johansen (1980) presented a solution in the context of general linear models and obtained the exact null distribution of the statistic.
Nel & Van der Merwe (1986) obtained the exact null distribution of the statistic
Kim (1992) presented another extension of the Welch approximate degrees-of-freedom solution by Yao (1965) but using the geometry of confidence ellipsoids for the mean vectors.
Krishnamoorthy & Yu (2004) proposed a new test based on a modification of the solution given by Nel & Van der Merwe (1986).
Gamage, Mathew, & Weerahandi (2004) proposed a procedure for testing equality of the mean vectors using the concept of generalized p-values which are functions of the sufficient statistics.
Yanagihara & Yuan (2005) proposed the F test, the Bartlett correction test and the modified Bartlett correction test.
Kawasaki & Seo (2015) propose an approximate solution to the problem by adjusting the degrees of freedom of the F distribution.
Multivariate statistical tests to compare two mean vectors
In this section we summarize the main multivariate statistical tests reported in the statistical literature for the multivariate Behrens-Fisher problem. For each test we show the statistic and its distribution under the null hypothesis. All tests reported here assume that the underlying distribution is the multivariate normal distribution.
The tests presented below need some inputs as the sample mean vectors
and the sample variance-covariance matrices
Next are listed the statistical tests to study the hypothesis:
Hotelling’s test
This test assumes that but unknow. The statistical test to perform Behrens-Fisher problem is
where the pooled variance-covariance matrix is given by:
If is true,
First order James’ test (1954)
This test was proposed by without assuming equality of the variance-covariance matrices. The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The critic value () for this test is given by
with
If then the null hypothesis given in Behrens-Fisher problem is rejected.
Yao’s test (1965)
This test was proposed by . The statistical test to perform Behrens-Fisher problem is
where the matrix is given by
If is true,
where is given by
with .
Johansen’s test (1980)
This test was proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
If is true,
where
NVM test (1986)
This test was proposed by without assuming equality of the variance-covariance matrices. The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
If is true,
with
Modified NVM test (2004)
This test was proposed by and it is a modification of the test proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
If is true,
with
Gamage’s test (2004)
This test was proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
If represents the observed value of , the generalized value for the test is where the is a random variable given by
To obtain the generalized value or , we should simulate at least 1000 values from each distribution , and independently, then those values are replaced in expression of T1 to obtain several , finally, the generalized value is obtained as the percentage rate .
In expression for T1 the values denoted by correspond to the eigenvalues of defined as
For more details about this to test please consult .
Yanagihara and Yuan’s test (2005)
This test was proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The null distribution for the statistic is
where the elements , and are defined as follows
Bartlett Correction test (2005)
This test was the second test proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The null distribution for the statistic is
where the elements and are defined as follows
Modified Bartlett Correction test (2005)
This test was the third test proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The null distribution for the statistic is
where the elements , and are defined as follows
Second Order Procedure (S procedure) test (2015)
This test was the first test proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The null distribution for the statistic is
where the elements and are defined as follows
The elements and are defined as:
Bias Correction Procedure (BC Procedure) test (2015)
This test was the second test proposed by . The statistical test to perform Behrens-Fisher problem is
where variance-covariance matrix is given by:
The null distribution for the statistic is
where the elements and are defined as follows
The elements are defined by the same expressions defined in the Second Order Procedure (S procedure) test whereas the elements and are given by
stests package
stests is an useful package in which are implemented several statistical tests for multivariate analysis. The current version of the package is hosted in github and any user could download the package using the next code.
if (!require('devtools')) install.packages('devtools')
devtools::install_github('fhernanb/stests', force=TRUE)To use the package we can use the usual way.
two_mean_vector_test is the main function in the
stests package to perform tests for the multivariate
Behrens-Fisher problem. The function deals with summarized data, this
means that the user should provide the sample means
,
,
the sample variances
,
,
and the sample sizes
and
.
two_mean_vector_test(xbar1, s1, n1, xbar2, s2, n2,
method="T2", alpha=0.05)In the next section, we present examples to illustrate the function use.
Examples
In this section we present some examples of the
two_mean_vector_test( ) function.
Hotelling’s test
The data correspond to the example 5.4.2 from page 137. The objective is to test the hypothesis versus .
Next are the input data to perform the test.
n1 <- 32
xbar1 <- c(15.97, 15.91, 27.19, 22.75)
s1 <- matrix(c(5.192, 4.545, 6.522, 5.25,
4.545, 13.18, 6.76, 6.266,
6.522, 6.76, 28.67, 14.47,
5.25, 6.266, 14.47, 16.65), ncol=4)
n2 <- 32
xbar2 <- c(12.34, 13.91, 16.66, 21.94)
s2 <- matrix(c(9.136, 7.549, 4.864, 4.151,
7.549, 18.6, 10.22, 5.446,
4.864, 10.22, 30.04, 13.49,
4.151, 5.446, 13.49, 28), ncol=4)To perform the test we use the two_mean_vector_test
function using method="T2" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="T2")
res
##
## T2 test for two mean vectors
##
## data: this test uses summarized data
## T2 = 97.678, F = 23.238, df1 = 4, df2 = 59, p-value = 1.444e-11
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 15.97 12.34
## xbar_2 15.91 13.91
## xbar_3 27.19 16.66
## xbar_4 22.75 21.94We can depict the value as follows.
plot(res, from=20, to=25, shade.col="tomato")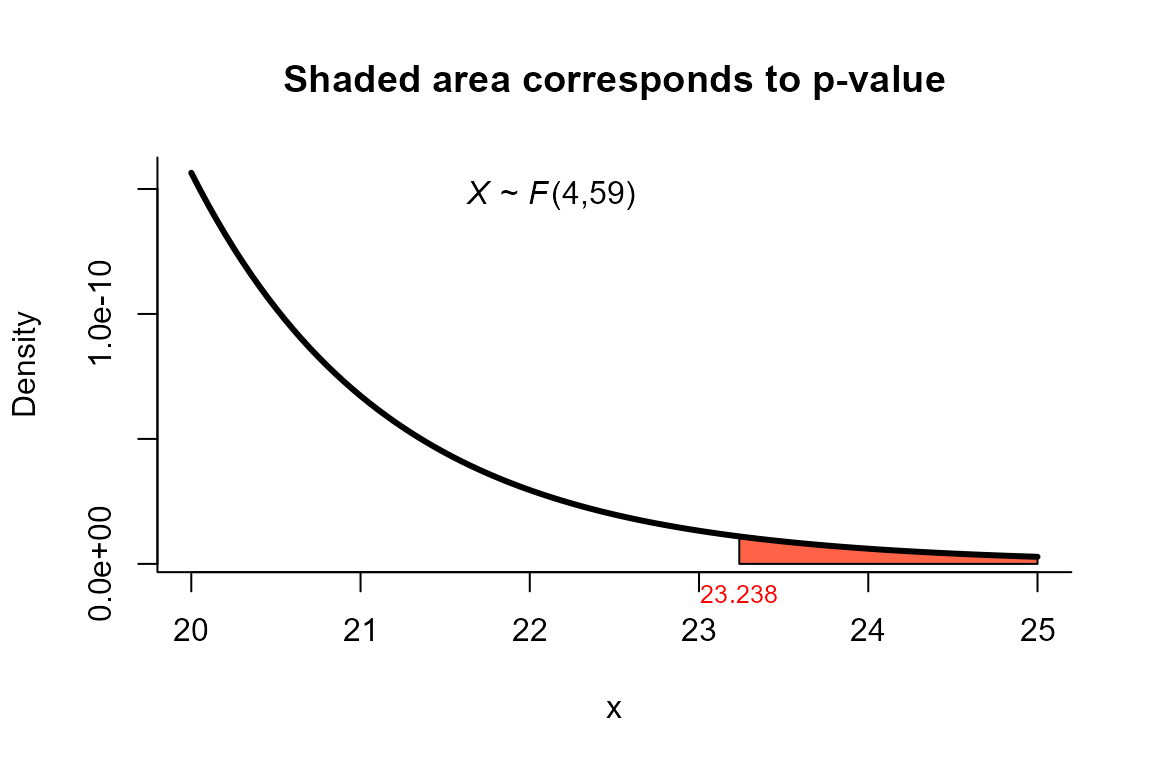
First order James’ test
The data correspond to the example in page 41 from . The objective is to test the hypothesis versus .
Next are the input data to perform the test.
n1 <- 16
xbar1 <- c(9.82, 15.06)
s1 <- matrix(c(120, -16.3,
-16.3, 17.8), ncol=2)
n2 <- 11
xbar2 <- c(13.05, 22.57)
s2 <- matrix(c(81.8, 32.1,
32.1, 53.8), ncol=2)To perform the test we use the two_mean_vector_test
function using method="james" with significance level
alpha=0.05 as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="james", alpha=0.05)
res
##
## James test for two mean vectors
##
## data: this test uses summarized data
## T2 = 9.4455, critic_value = 7.2308, df = 2
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 9.82 13.05
## xbar_2 15.06 22.57We can depict the value as follows.
plot(res, from=5, to=10, shade.col="lightgreen")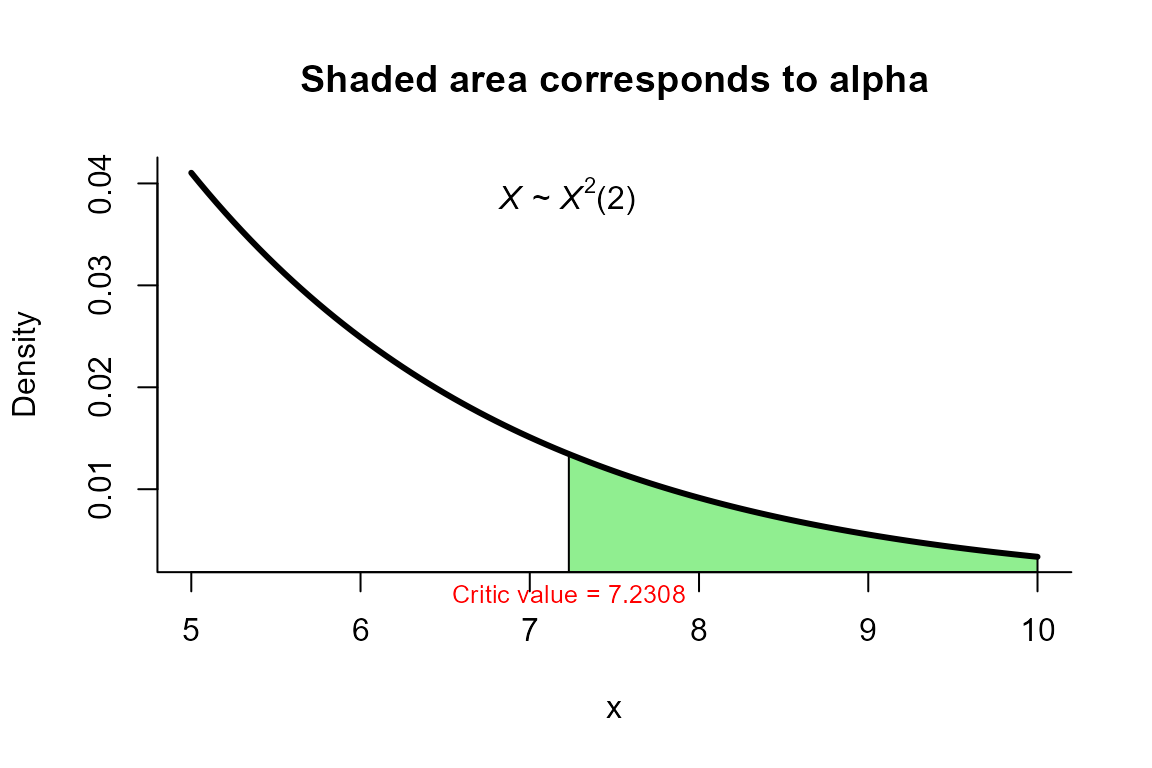
Yao’s test
The data correspond to the example in page 141 from . The objective is to test the hypothesis versus .
Next are the input data to perform the test.
n1 <- 16
xbar1 <- c(9.82, 15.06)
s1 <- matrix(c(120, -16.3,
-16.3, 17.8), ncol=2)
n2 <- 11
xbar2 <- c(13.05, 22.57)
s2 <- matrix(c(81.8, 32.1,
32.1, 53.8), ncol=2)To perform the test we use the two_mean_vector_test
function using method="yao" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="yao")
res
##
## Yao test for two mean vectors
##
## data: this test uses summarized data
## T2 = 9.4455, F = 4.3855, df1 = 2.000, df2 = 13.001, p-value = 0.03503
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 9.82 13.05
## xbar_2 15.06 22.57We can depict the value as follows.
plot(res, from=2, to=6, shade.col="pink")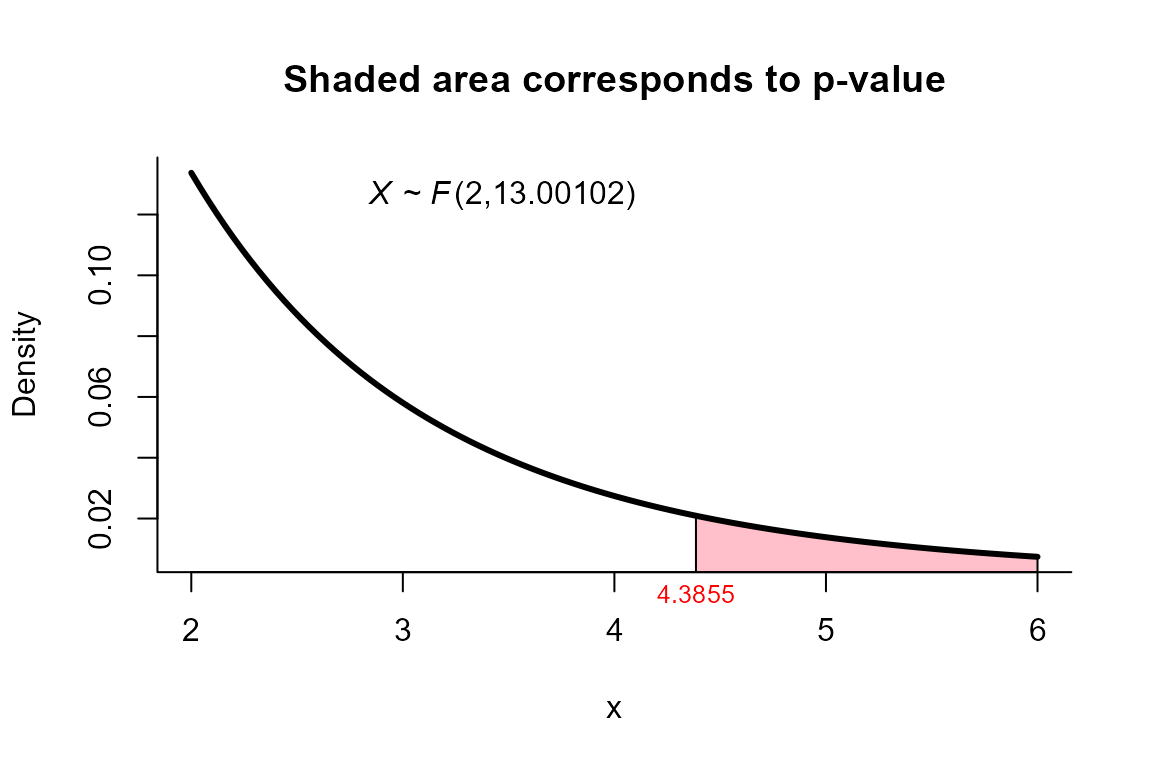
Johansen’s test
For this example we use the same dataset described in the example for
Yao’s test but using the method="johansen" as follows.
Next are the input data to perform the test.
n1 <- 16
xbar1 <- c(9.82, 15.06)
s1 <- matrix(c(120, -16.3,
-16.3, 17.8), ncol=2)
n2 <- 11
xbar2 <- c(13.05, 22.57)
s2 <- matrix(c(81.8, 32.1,
32.1, 53.8), ncol=2)To perform the test we use the two_mean_vector_test
function using method="johansen" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="johansen")
res
##
## Johansen test for two mean vectors
##
## data: this test uses summarized data
## T2 = 9.4455, F = 4.5476, df1 = 2.000, df2 = 16.309, p-value = 0.02587
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 9.82 13.05
## xbar_2 15.06 22.57We can depict the value as follows.
plot(res, from=2, to=6, shade.col="aquamarine1")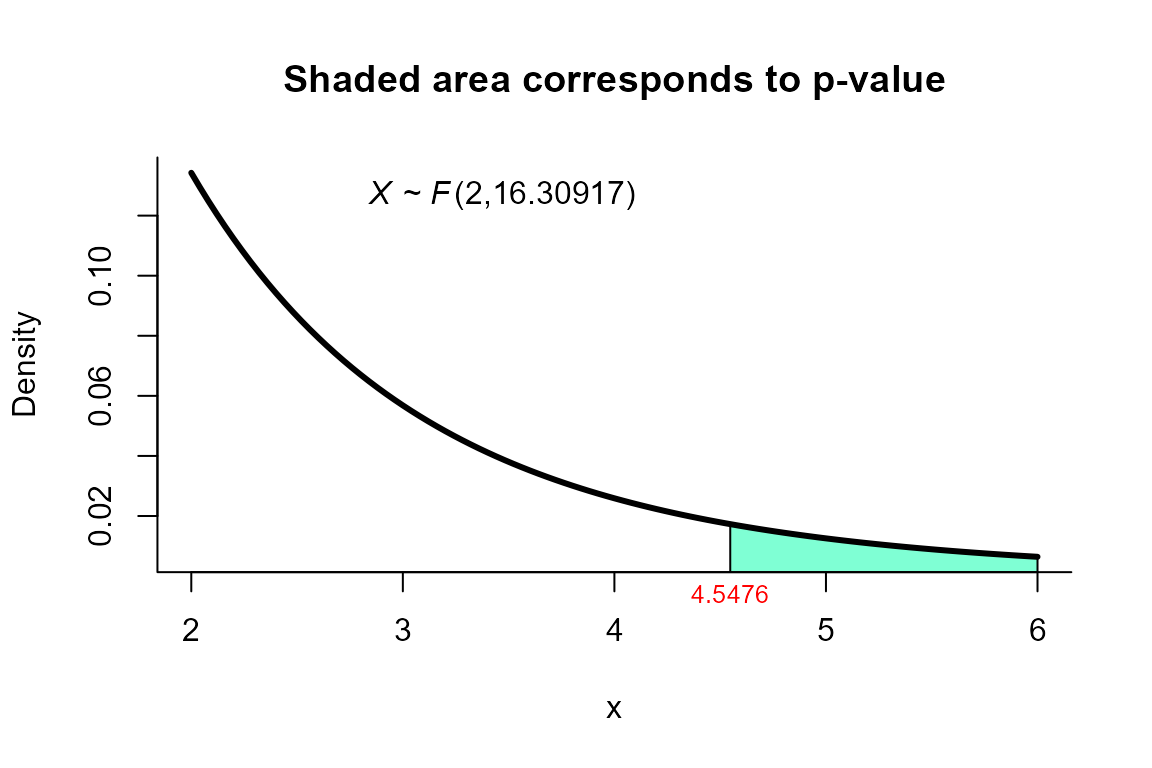
NVM test
The data correspond to the example 4.1 from page 3729. The objective is to test the hypothesis versus .
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="nvm" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="nvm")
res
##
## Nel and Van der Merwe test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.6585, F = 7.7402, df1 = 2.000, df2 = 86.873, p-value =
## 0.0008064
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=6, to=10, shade.col="cyan2")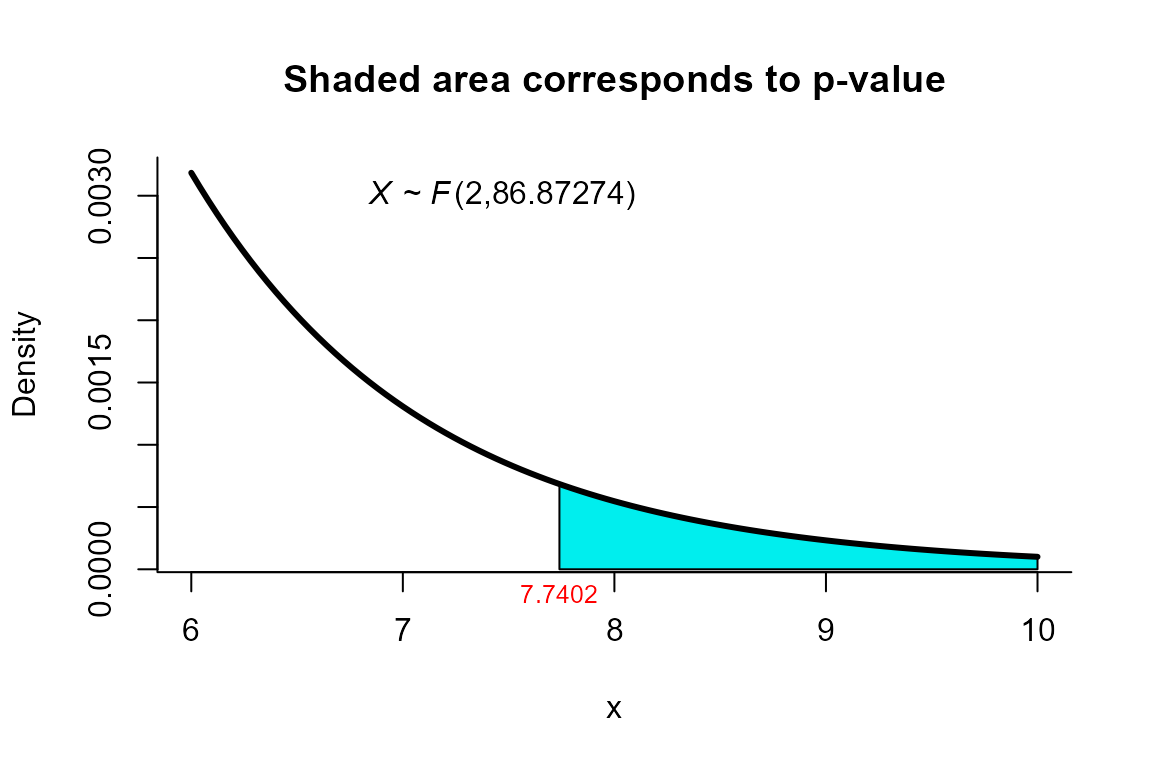
Modified NVM test
For this example we use the same dataset described in the example for
NVM test but using the method="mnvn" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="mnvm" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="mnvm")
res
##
## Modified Nel and Van der Merwe test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.6585, F = 7.7261, df1 = 2.000, df2 = 74.906, p-value = 0.00089
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=7, to=8, shade.col="lightgoldenrodyellow")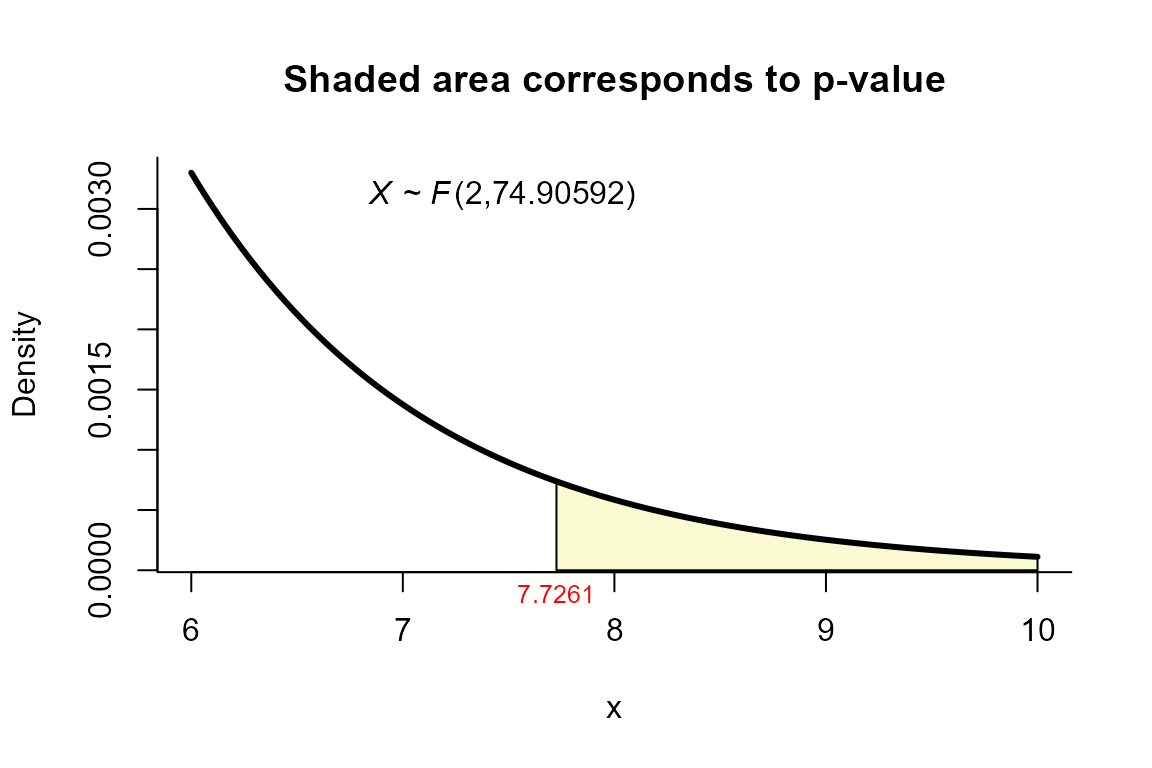
Gamage’s test
For this example we use the same dataset described in the example for
NVM test but using the method="gamage" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="gamage" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="gamage")
res
##
## Gamage test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.659, p-value = 0.0015
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=0, to=20, shade.col="lightgoldenrodyellow")
Yanagihara and Yuan’s test
For this example we use the same dataset described in the example for
NVM test but using the method="yy" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="yy" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="yy")
res
##
## Yanagihara and Yuan test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.6585, F = 7.7272, df1 = 2.000, df2 = 74.309, p-value =
## 0.0008937
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=6, to=10, shade.col='gold2')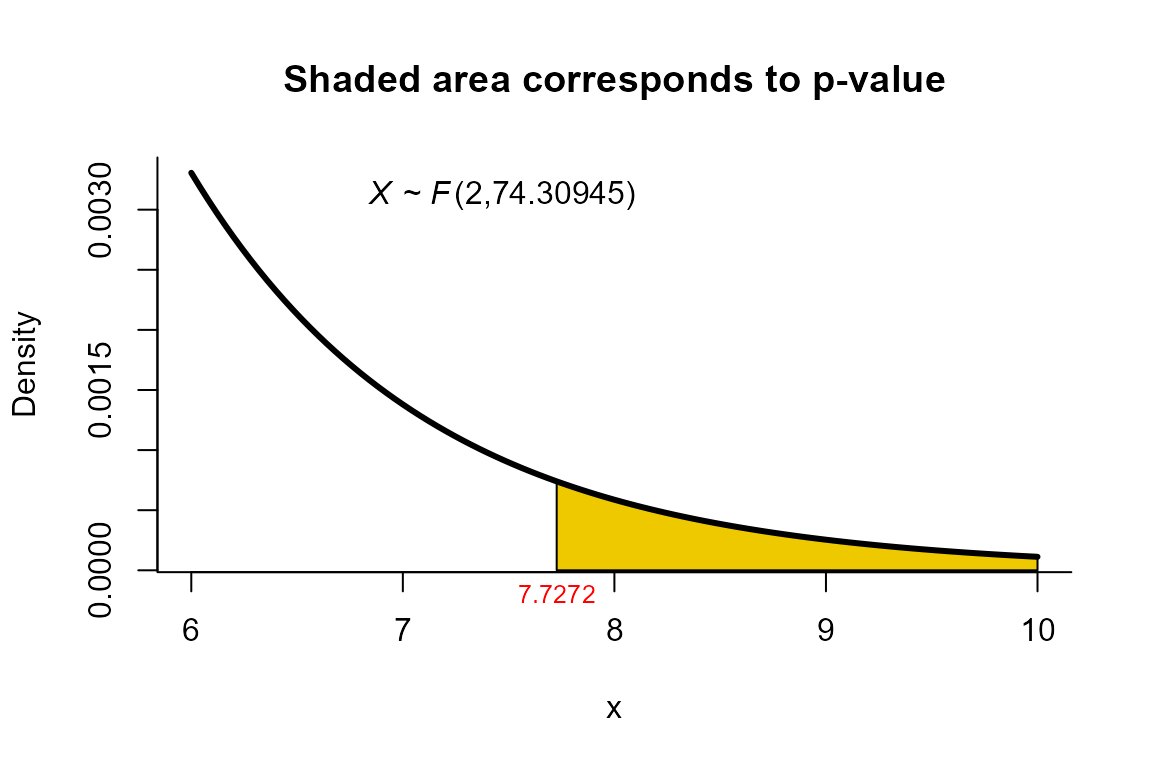
Bartlett Correction test
For this example we use the same dataset described in the example for
NVM test but using the method="byy" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="byy" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="byy")
res
##
## Bartlett Correction test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.659, X-squared = 15.040, df = 2, p-value = 0.0005422
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=12, to=20, shade.col='hotpink2')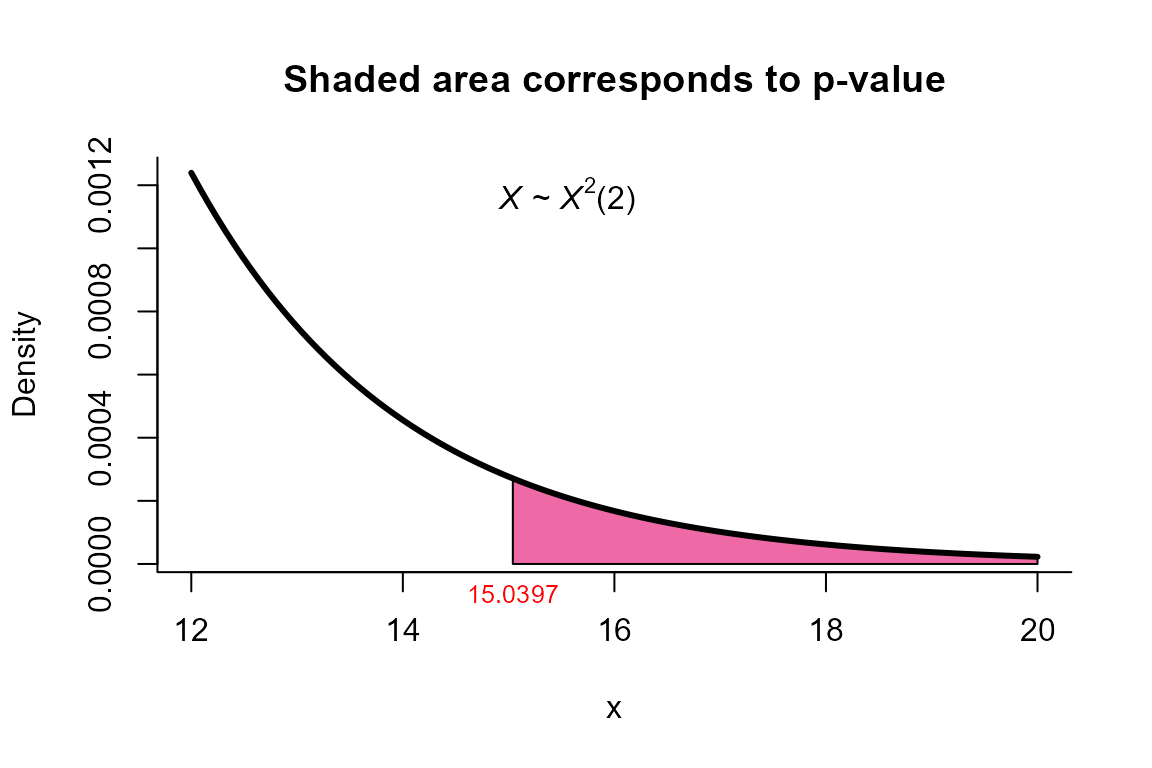
Modified Bartlett Correction test
For this example we use the same dataset described in the example for
NVM test but using the method="mbyy" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="mbyy" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="mbyy")
res
##
## Modified Bartlett Correction test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.659, X-squared = 14.044, df = 2, p-value = 0.000892
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=12, to=20, shade.col='green3')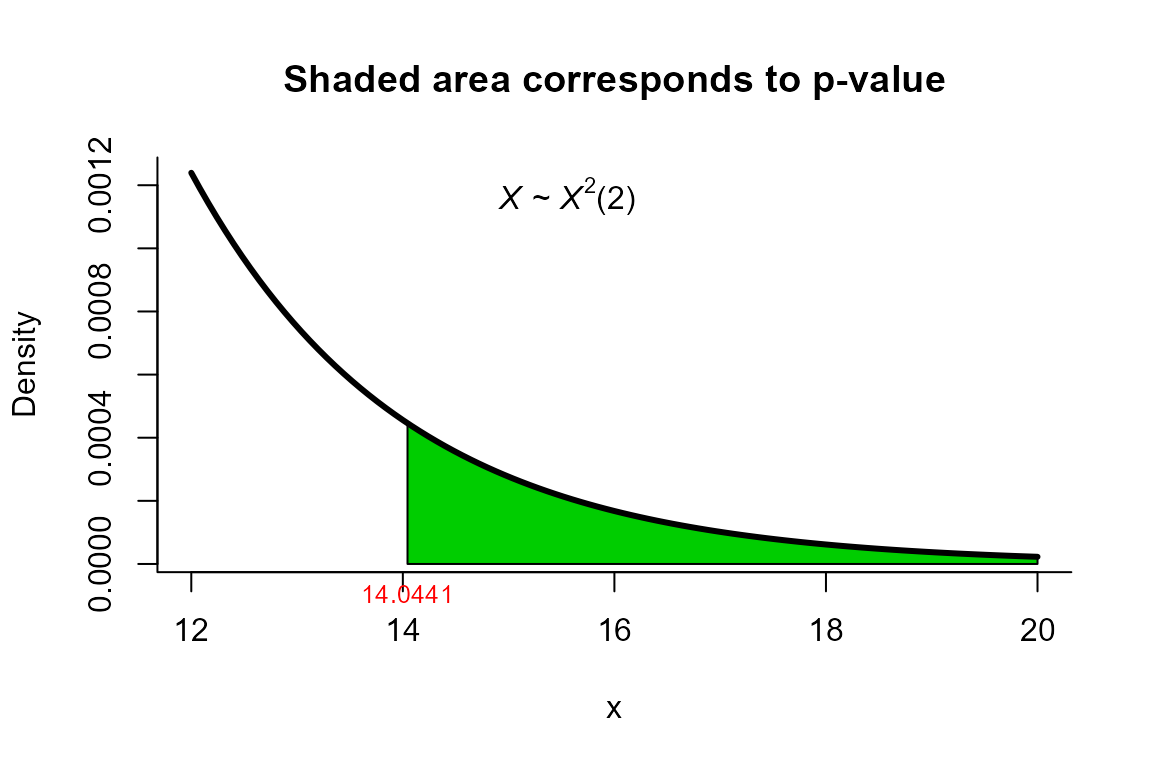
Second Order Procedure (S procedure) test
For this example we use the same dataset described in the example for
NVM test but using the method="ks1" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="ks1" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="ks1")
res
##
## Kawasaki and Seo (Second order) test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.6585, F = 7.7247, df1 = 2.000, df2 = 49.576, p-value = 0.001201
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=6, to=10, shade.col='hotpink1')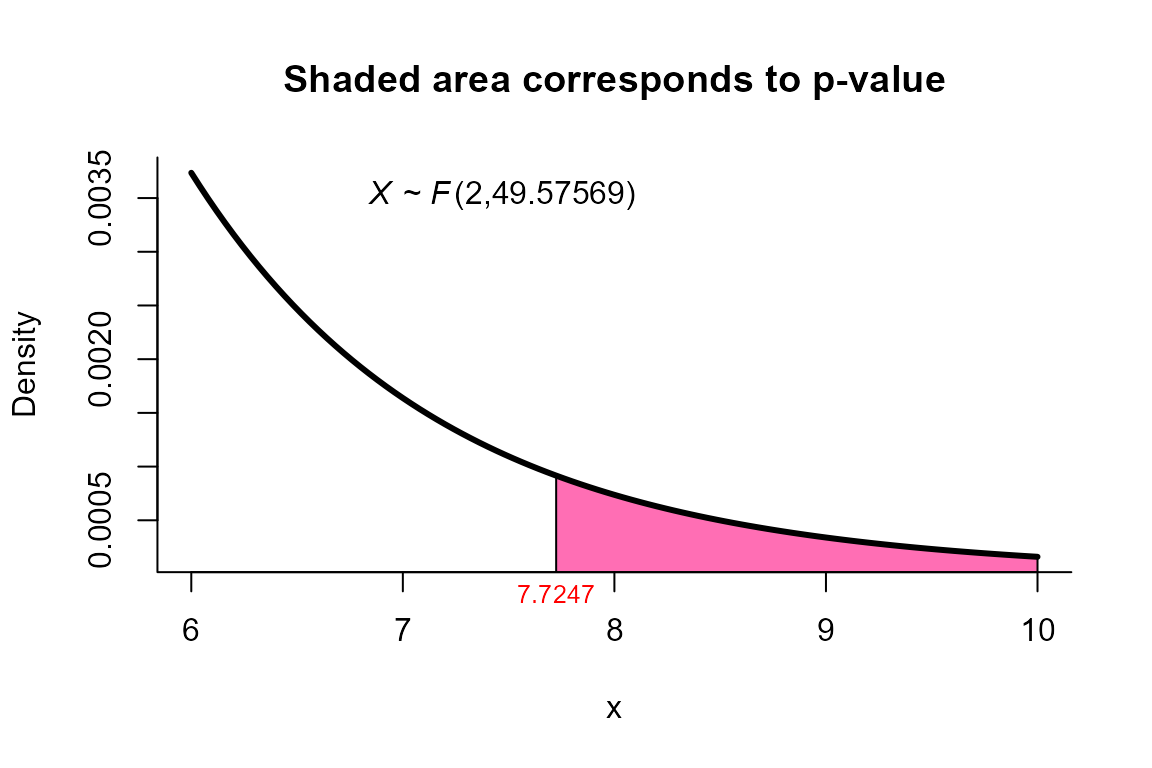
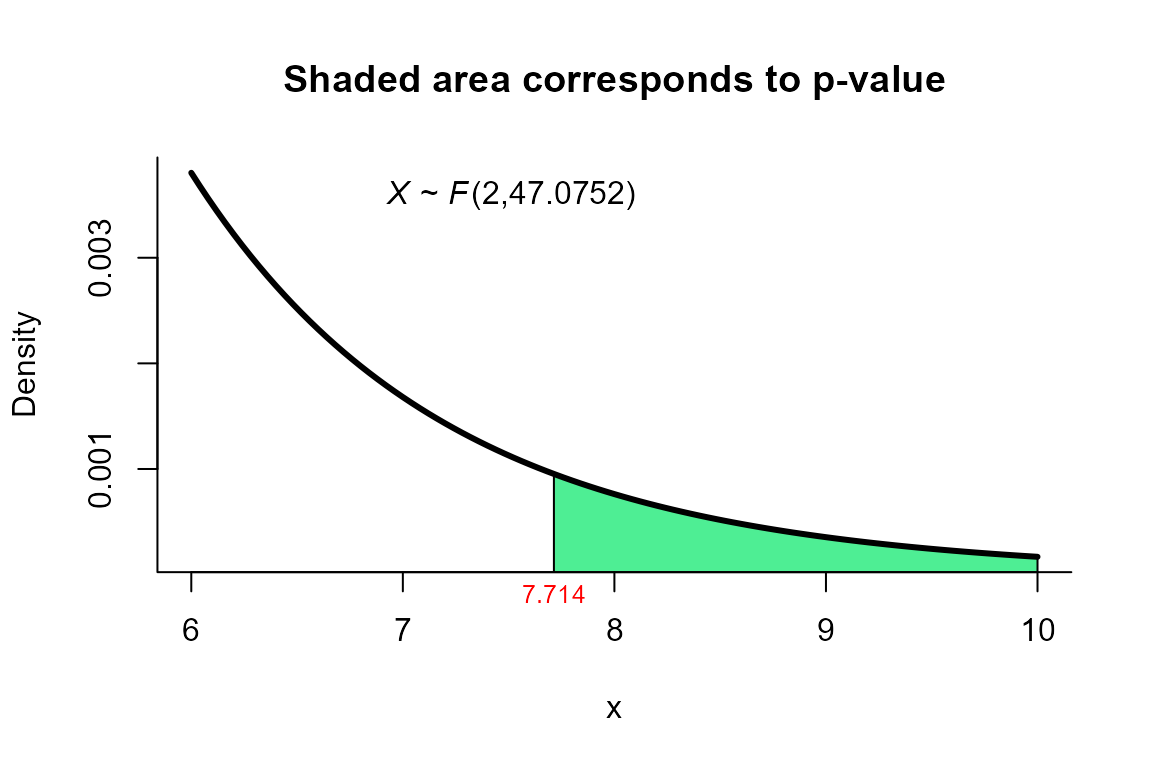
Bias Correction Procedure (BC Procedure) test
For this example we use the same dataset described in the example for
NVM test but using the method="ks2" as follows.
Next are the input data to perform the test.
n1 <- 45
xbar1 <- c(204.4, 556.6)
s1 <- matrix(c(13825.3, 23823.4,
23823.4, 73107.4), ncol=2)
n2 <- 55
xbar2 <- c(130.0, 355.0)
s2 <- matrix(c(8632.0, 19616.7,
19616.7, 55964.5), ncol=2)To perform the test we use the two_mean_vector_test
function using method="ks2" as follows.
res <- two_mean_vector_test(xbar1=xbar1, s1=s1, n1=n1,
xbar2=xbar2, s2=s2, n2=n2,
method="ks2")
res
##
## Kawasaki and Seo (Bias Correction) test for two mean vectors
##
## data: this test uses summarized data
## T2 = 15.659, F = 7.714, df1 = 2.000, df2 = 47.075, p-value = 0.001266
## alternative hypothesis: mu1 is not equal to mu2
##
## sample estimates:
## Sample 1 Sample 2
## xbar_1 204.4 130
## xbar_2 556.6 355We can depict the value as follows.
plot(res, from=6, to=10, shade.col='seagreen2')