Tests for homogeneity of covariances matrices
Source:R/mult_var_matrices_test.R
mult_var_matrices_test.RdThe function implements the test for \(H_0: \Sigma_1 = \Sigma_2 = ... = \Sigma_g\) versus \(H_1\) at least one \(\Sigma_i\) is different.
mult_var_matrices_test(S, N, method = "box")Arguments
Value
A list with class "htest" containing the following components:
- statistic
the value of the statistic.
- parameter
the degrees of freedom for the test.
- p.value
the p-value for the test.
- estimate
the estimated mean vectors.
- method
a character string indicating the type of test performed.
Details
the "method" must be one of "box" (default),
"modified_LRT", "wald_schott".
To know in detail all tests implemented here the reader can visit the vignette https://fhernanb.github.io/stests/articles/Tests_Sigmas.html
References
Schott, J. R. (2001). Some tests for the equality of covariance matrices. Journal of statistical planning and inference, 94(1), 25-36.
Schott, J. R. (2007). A test for the equality of covariance matrices when the dimension is large relative to the sample sizes. Computational Statistics & Data Analysis, 51(12), 6535-6542.
Mardia, K. V., Kent, J. T., & Bibby, J. M. (1979). Multivariate analysis.
Examples
# Example 5.2.3 from Diaz and Morales (2015) page 200
S1 <- matrix(c(12.65, -16.45,
-16.45, 73.04), ncol=2, nrow=2)
S2 <- matrix(c(11.44, -27.77,
-27.77, 100.64), ncol=2, nrow=2)
S3 <- matrix(c(14.46, -31.26,
-31.26, 101.03), ncol=2, nrow=2)
N1 <- 26
N2 <- 23
N3 <- 25
S <- list(S1, S2, S3)
N <- list(N1, N2, N3)
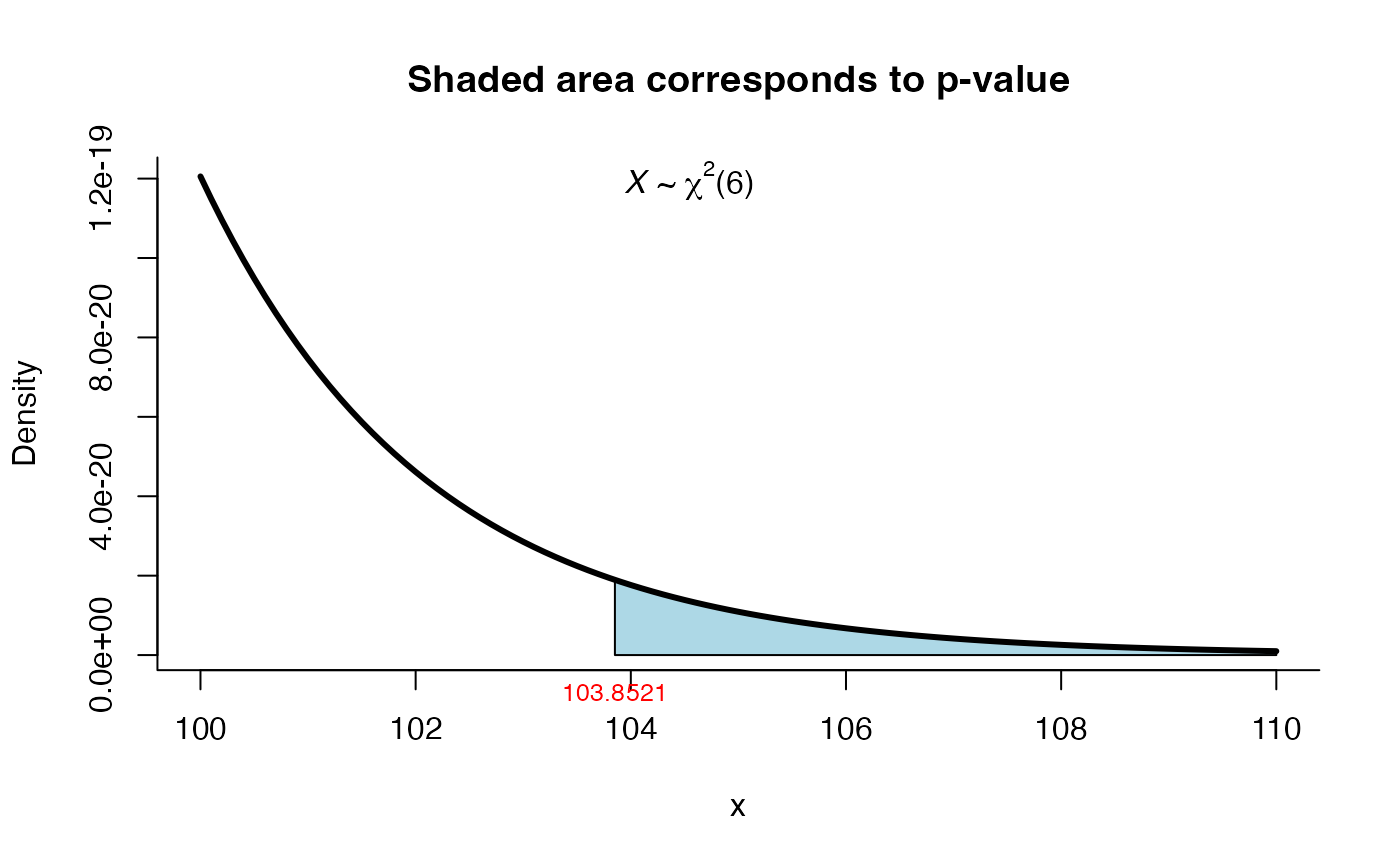
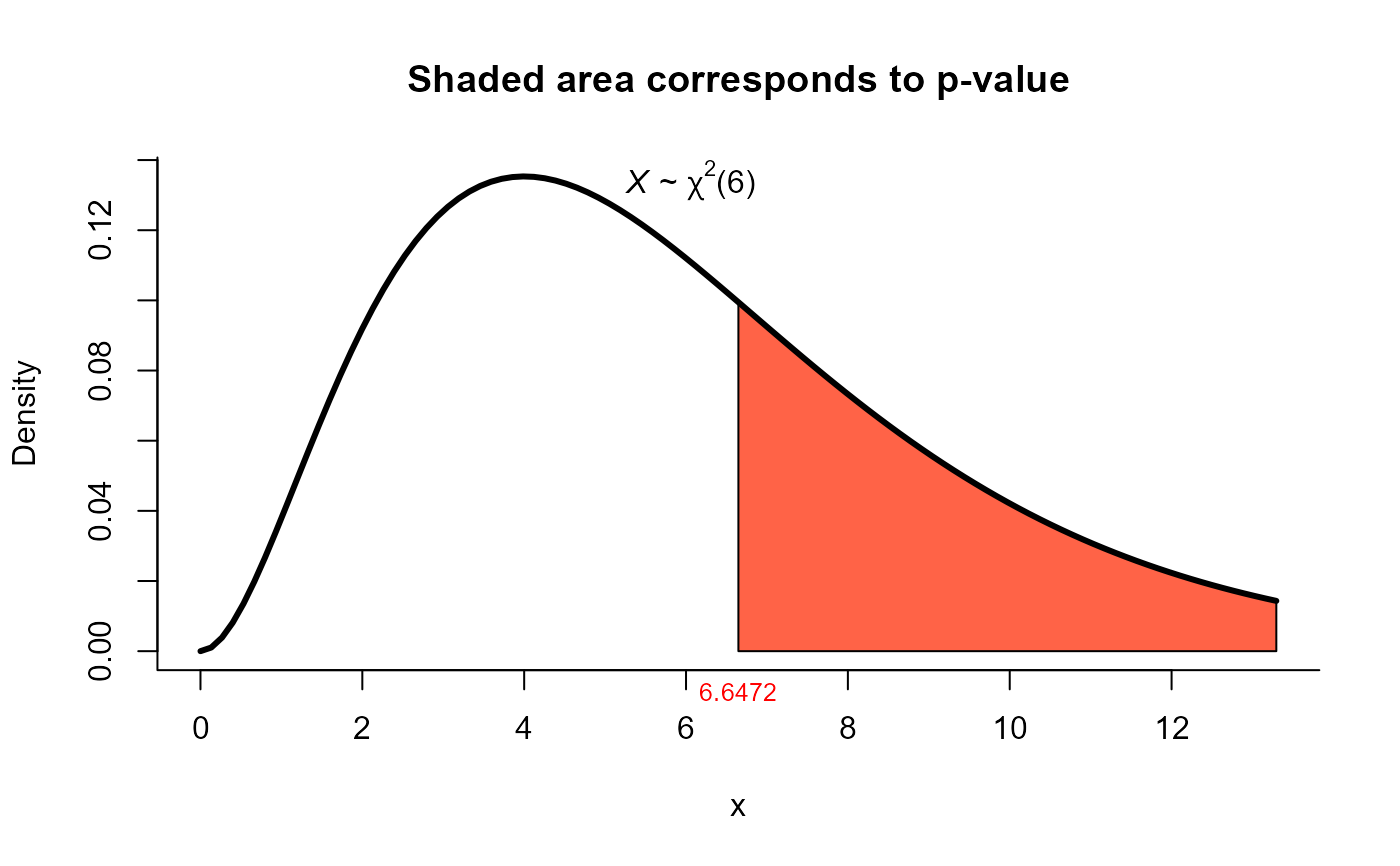
res <- mult_var_matrices_test(S=S, N=N, method="box")
res
#>
#> Box test for homogeneity of covariances
#>
#> data: this test uses summarized data
#> phi = 6.6472, df = 6, p-value = 0.3547
#> alternative hypothesis: at least one covariance matrix is different
#>
#> sample estimates:
#> [1] "Due to the high value of m, matrices S1, ..., S3 are not displayed."
#>
plot(res, shade.col="tomato")
 # Example 5.3.4 from Mardia (1979) page 141
S1 <- matrix(c(132.99, 75.85, 35.82,
75.85, 47.96, 20.75,
35.82, 20.75, 10.79), ncol=3, nrow=3)
S2 <- matrix(c(432.58, 259.87, 161.67,
259.87, 164.57, 98.99,
161.67, 98.99, 63.87), ncol=3, nrow=3)
N1 <- 24
N2 <- 24
S <- list(S1, S2)
N <- list(N1, N2)
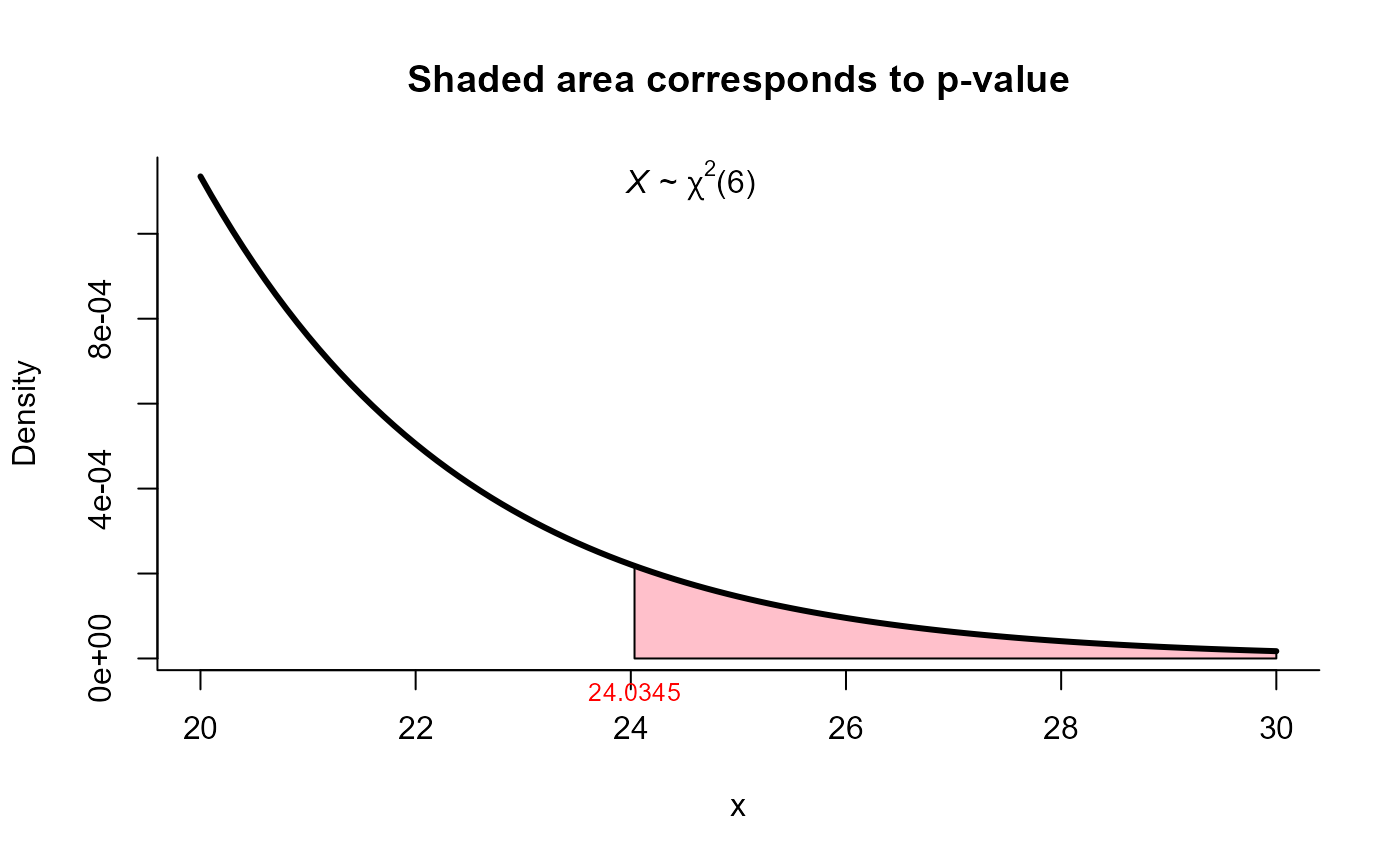
res <- mult_var_matrices_test(S=S, N=N, method="modified_LRT")
res
#>
#> Modified likelihood ratio (or Bartlett's) test for homogeneity of
#> covariances
#>
#> data: this test uses summarized data
#> M = 25.862, df = 6, p-value = 0.0002362
#> alternative hypothesis: at least one covariance matrix is different
#>
#> sample estimates:
#> [1] "Due to the high value of m, matrices S1, ..., S2 are not displayed."
#>
plot(res, from=20, to=30, shade.col="pink")
# Example 5.3.4 from Mardia (1979) page 141
S1 <- matrix(c(132.99, 75.85, 35.82,
75.85, 47.96, 20.75,
35.82, 20.75, 10.79), ncol=3, nrow=3)
S2 <- matrix(c(432.58, 259.87, 161.67,
259.87, 164.57, 98.99,
161.67, 98.99, 63.87), ncol=3, nrow=3)
N1 <- 24
N2 <- 24
S <- list(S1, S2)
N <- list(N1, N2)
res <- mult_var_matrices_test(S=S, N=N, method="modified_LRT")
res
#>
#> Modified likelihood ratio (or Bartlett's) test for homogeneity of
#> covariances
#>
#> data: this test uses summarized data
#> M = 25.862, df = 6, p-value = 0.0002362
#> alternative hypothesis: at least one covariance matrix is different
#>
#> sample estimates:
#> [1] "Due to the high value of m, matrices S1, ..., S2 are not displayed."
#>
plot(res, from=20, to=30, shade.col="pink")
 res <- mult_var_matrices_test(S=S, N=N, method="wald_schott")
res
#>
#> Wald-Schott test for homogeneity of covariances
#>
#> data: this test uses summarized data
#> W = 103.85, df = 6, p-value < 2.2e-16
#> alternative hypothesis: the covariance matrices are different
#>
#> sample estimates:
#> [1] "Due to the high value of m, matrices S1, ..., S2 are not displayed."
#>
plot(res, from=100, to=110, shade.col="lightblue")
res <- mult_var_matrices_test(S=S, N=N, method="wald_schott")
res
#>
#> Wald-Schott test for homogeneity of covariances
#>
#> data: this test uses summarized data
#> W = 103.85, df = 6, p-value < 2.2e-16
#> alternative hypothesis: the covariance matrices are different
#>
#> sample estimates:
#> [1] "Due to the high value of m, matrices S1, ..., S2 are not displayed."
#>
plot(res, from=100, to=110, shade.col="lightblue")